Impelementing XOR with NEAT
Preface
All the executions and plots are generated from scratch at build time. This means results may vary from execution to execution.
This also creates some constraints in terms of execution time. A proper analysis would have us execute each approach many times prior to making any generalization, which we cannot afford here.
While I could use the same seed for the random generator and therefore generate reproducible results, this wouldn’t be befitting of a demonstration of evolutionary algorithms.
Note also the purpose of this document is to serve me as notes and is not meant to be taken as exhaustive or a fully fledged blog post.
Introduction
Implementing a XOR gate with NEAT is a typical demonstration as it is not only used in the original paper Evolving Neural Networks Through Augmenting Topologies but also a problem that does require the use of an additional node in the network.
Therefore it is an ideal use case to explore.
Methodology
Chromosomes
We want to implement a XOR gate, and as such will need 2 input nodes and 1 output node. We will also add a bias node that will always be set to 1. The networks will be fully connected upon initialization and look something like:
The connections will be enabled and their weight randomly selected.
The network used will be a feed forward network (ie. no loops), and will use tanh as its activation function.
This translates in code to:
final var feedForwardNetwork = new FeedForwardNetwork(inputNodeIndices,
outputNodeIndices,
connections,
Activations.tanhFloat);Where connections is the list of connections defining the network
Fitness
The fitness will be computed by evaluating each of the possible inputs and comparing them with the expected output of a XOR gate. The closer to the expectations, the higher will the score be.
Since we want a more fine grained fitness for better comparing individuals, we will use the distance to the expectations as an error measurement and deduct it from the ideal score. Thus scores can go from 0 to 4, where 4 means a perfect score.
The code to implement the fitness is:
final int outputNodeIndex = outputNodeIndices.iterator().next();
for (final boolean a : Set.of(false, true)) {
final float aValue = a ? 1 : 0;
for (final boolean b : Set.of(false, true)) {
final float bValue = b ? 1 : 0;
final float expectedOutput = a ^ b ? 1 : 0;
// node 2 is bias
final var inputValues = Map.of(0, aValue, 1, bValue, 2, 1.0f);
final Map<Integer, Float> computedOutputValues = feedForwardNetwork.compute(inputValues);
final float computedOutput = computedOutputValues.get(outputNodeIndex);
if (print) {
logger.info(
"a: {}, b: {} ===> {} ({}) / {}",
aValue,
bValue,
computedOutput,
outputNodeIndex,
expectedOutput);
}
errorDistance += Math.sqrt((expectedOutput - computedOutput) * (expectedOutput - computedOutput));
}
}
return 4 - errorDistance;Configuration
To specify the problem, we can use a standard EAConfiguration with the following tweaks:
- We define the chromosome of type NeatChromosome with 3 inputs and one output
- We have a specific selection for the parents
- We have a specific set of operators for the mutations
- We have a specific NeatCombination that implements the combination of two parents as specified in the NEAT paper
- We assume a fitness equal or greater to 3.95 is good enough to our goal
The configuration is defined as:
final Builder<Float> eaConfigurationBuilder = new EAConfiguration.Builder<>();
eaConfigurationBuilder.chromosomeSpecs(NeatChromosomeSpec.of(3, 1, -5, 5))
.parentSelectionPolicy(neatSelection)
.combinationPolicy(NeatCombination.build())
.mutationPolicies(neatMutations)
.fitness(fitnessNEAT(false))
.optimization(Optimization.MAXIMIZE)
.termination(
Terminations.<Float>or(Terminations.ofStableFitness(300), Terminations.ofFitnessAtLeast(3.95f)));With regards to the selection policy, while there exist a factory with sensible defaults (see NeatSelection::ofDefault ), we do want to ensure the following:
- We only keep the top 20% of the individuals for each species
- We will always keep at least 1 individual for a species
- Selecting individuals within a species is done through tournaments
Thus the code will be:
final SelectionPolicy neatSelection = NeatSelection.builder()
.minSpeciesSize(1)
.perSpeciesKeepRatio(.20f)
.speciesSelection(Tournament.of(3))
.speciesPredicate(
(i1, i2) -> NeatUtils.compatibilityDistance(i1.genotype(), i2.genotype(), 0, 2, 2, 1f) < 4.5)
.build();In terms of mutations, we can find the two main types of mutations being separated:
- The mutations that affect weights, such as creep, neat or random changes.
- The mutations that affect the structure
It is to be noted how high are set the mutations:
final List<MutationPolicy> neatMutations = List.of(
MultiMutations.of(
CreepMutation.of(0.85f, NormalDistribution.of(0.0, 0.333)),
RandomMutation.of(0.85f),
NeatConnectionWeight.builder().populationMutationProbability(0.85f).build()),
MultiMutations.of(
SwitchStateMutation.of(0.01f),
AddNode.of(0.20f),
DeleteNode.of(0.20f),
AddConnection.of(0.5f),
DeleteConnection.of(0.5f)));And then we need the execution context, which will be very simple as we only want to specify the population and some evolution listeners so we can log and record how the algorithm performs:
final var eaExecutionContextBuilder = NeatEAExecutionContexts.<Float>standard();
eaExecutionContextBuilder.populationSize(populationSize).evolutionListeners(evolutionListeners);
final var eaExecutionContext = eaExecutionContextBuilder.build();Finally, we can launch the computations:
final EASystem<Float> eaSystem = EASystemFactory.from(eaConfiguration, eaExecutionContext);
final EvolutionResult<Float> evolutionResult = eaSystem.evolve();
logger.info("Best genotype: {}", evolutionResult.bestGenotype());
logger.info("\twith fitness: {}", evolutionResult.bestFitness());
logger.info("\tat generation: {}", evolutionResult.generation());Results
The results are pretty nice and it will consistently find a solution very quickly. The networks are small and do tend to exhibit similar patterns.
It is interesting to note that the bias node has consistently its connections disabled or outright removed.
Here is an example of solution:
Note: remember that all the results in this article are re-computed at each build.
We can also look at the evolution of the fitness over time:
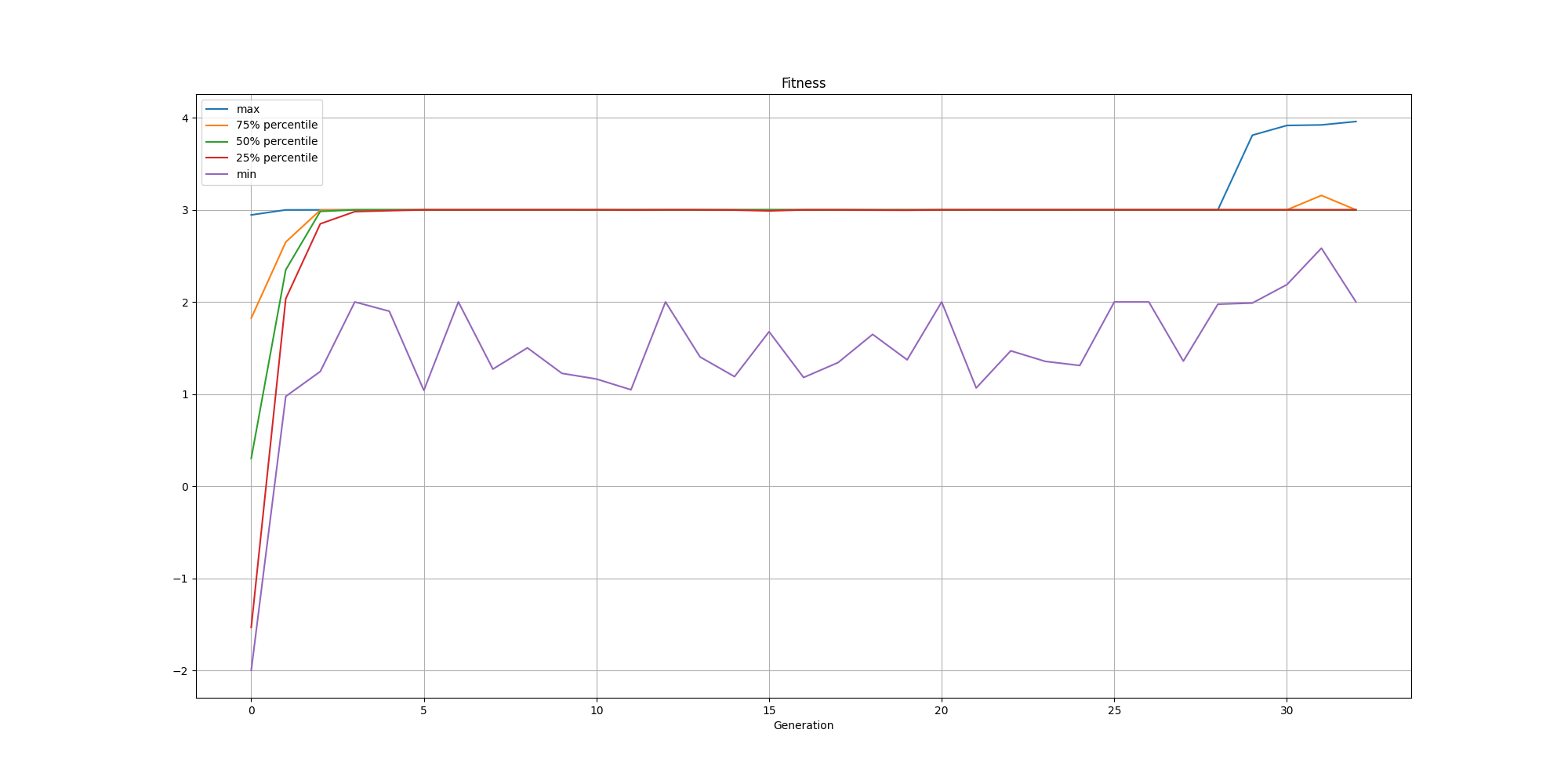
It is also interesting to look at the nodes:
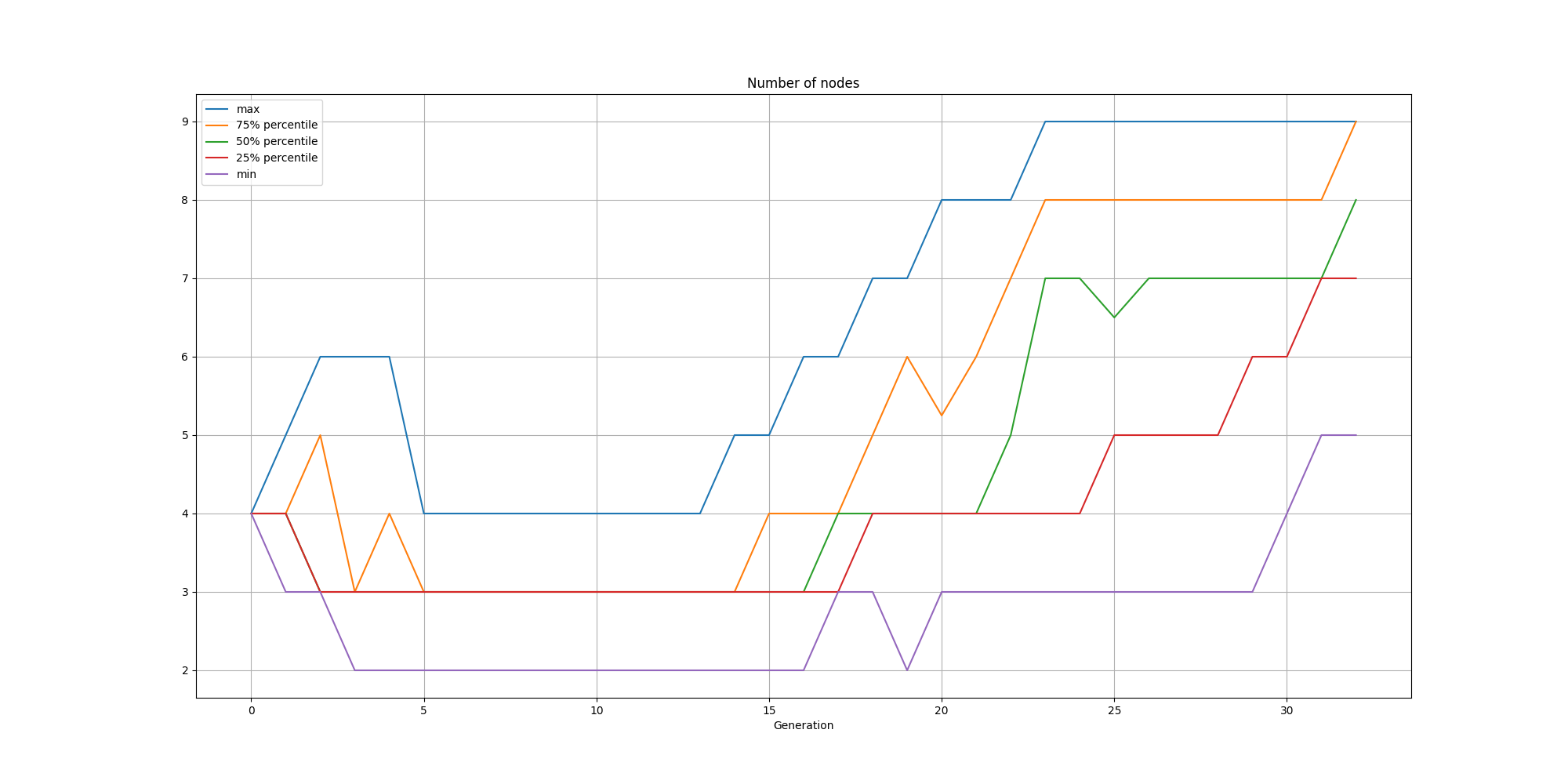
And the connections:
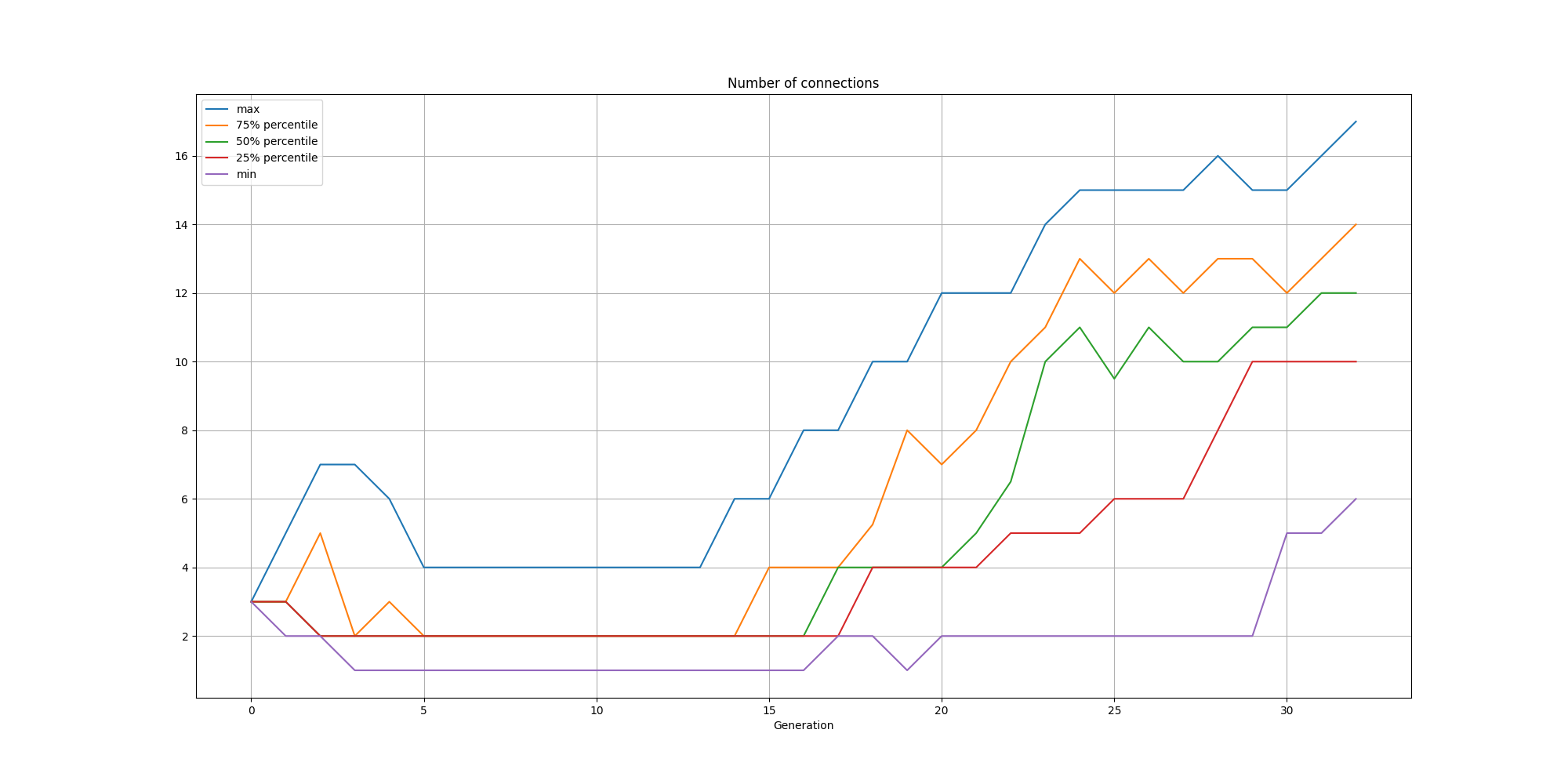
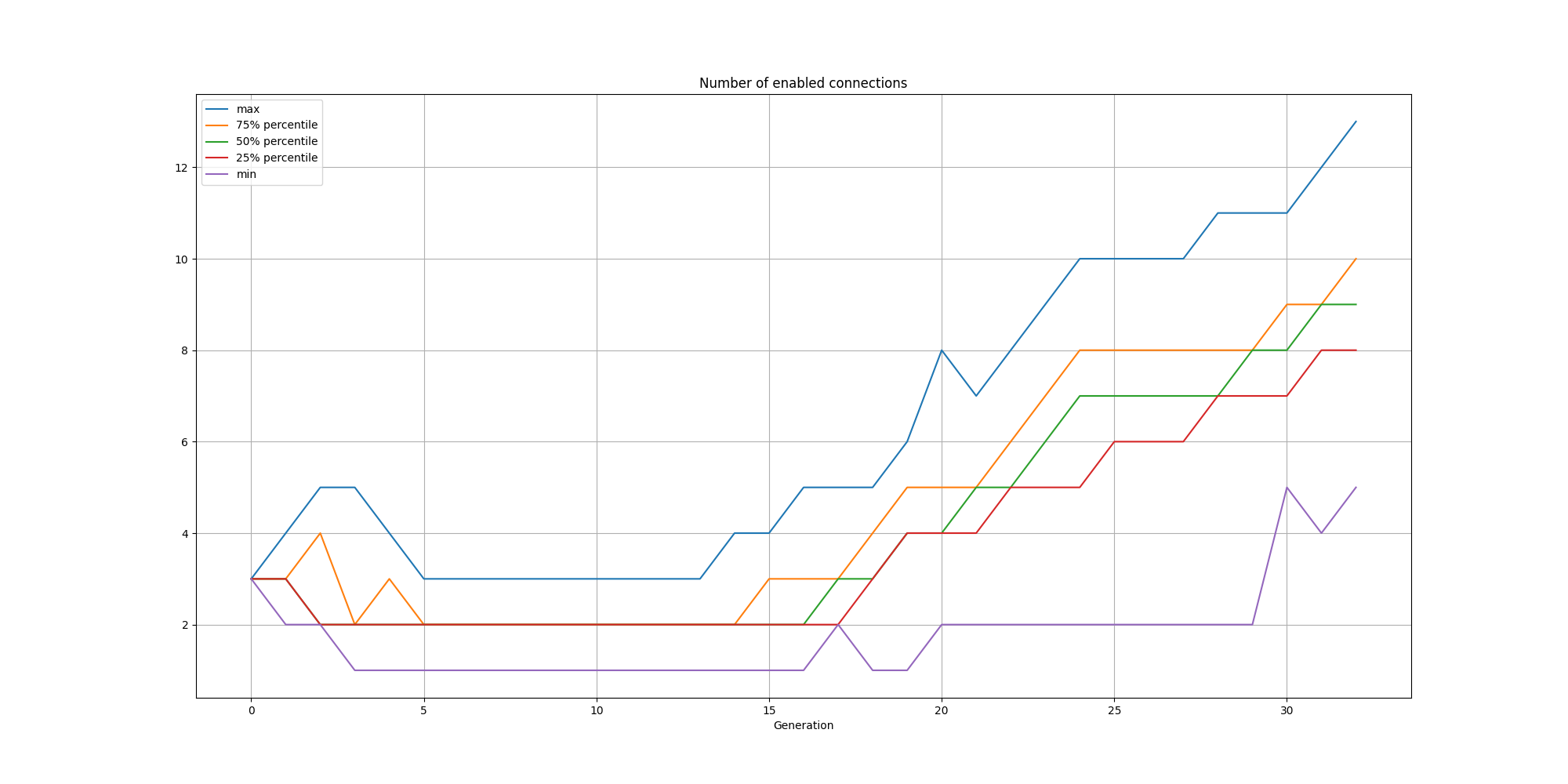
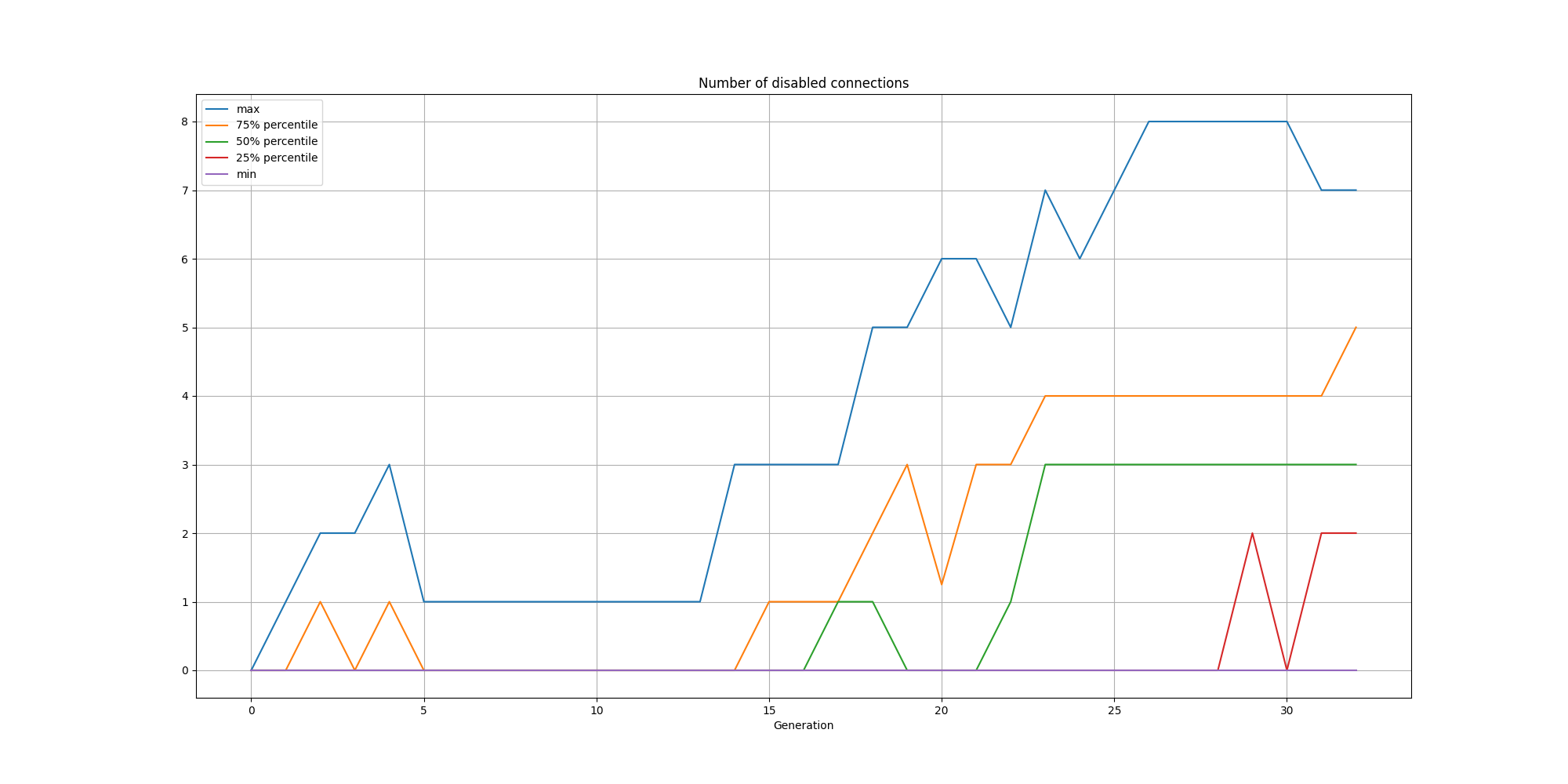
We can observe that while the solutions are consistently small, there is a steady increase in nodes and connections over time.
Conclusion
NEAT is a very powerful algorithm that can derive great results very quickly. However its parameters are very sensitive and could lead to a different outcome if not tweak appropriately.
There are also a few topics I would like to dive deeper into in the future:
- Visualizing the species over time. It would be nice to see how they evolve over time
- Digging into the relationship between number of nodes and connection with their fitness
- Digging into the sensibitility to parameters
- Changing starting conditions and not using a fully connected network
- Automatically adjust the compatibility threshold as discussed in the NEAT FAQ